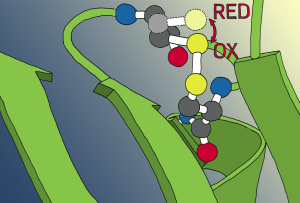
In the presence of oxidative and nitrosative stress proteins can undergo oxidative modification. While unintended oxidative modifications most often lead to the damage of the affected proteins, oxidative modifications also play important roles in cellular redox sensing. Classical redox sensor proteins use reversible modifications of cysteine to change their activity in response to a changing redox environment. These redox sensors are involved in a wide variety of cellular processes ranging from central energy metabolism over protein quality control to the regulation of the oxidative stress response.
When bacteria are phagocytized by white cells, they are exposed to a toxic cocktail of oxidants that leads to a breakdown of their intracellular redox state. Successful pathogenic bacteria must be able to protect themselves from those toxic oxidants. E. coli has a number of proteins that help this bacterium to overcome oxidative stress. Some of them are activated by cysteine oxidation, others by N-chlorination of basic amino acids. We study the role of these post-translational modifications with redox proteomics, biochemistry, molecular biology, genetics and bioinformatics.
The host also suffers from the oxidative stress initiated by its own immune system. Our lab has discovered proteins in our blood, which change their function when exposed to the oxidant hypochlorous acid and can protect and stimulate immune cells.
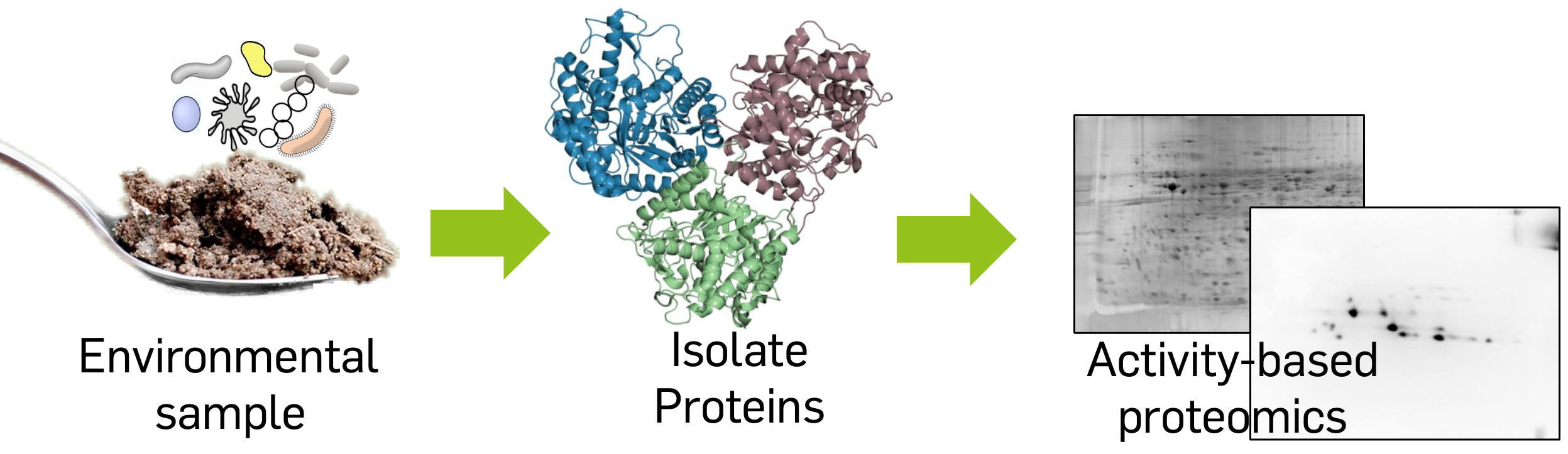
Bacteria can metabolize a wide variety of substrates, equipping them with a plethora of biocatalysts. Our lab is using a functional metaproteomics approach to find new biocatalysts in environmental samples. Combining the immediacy of traditional activity-based screening with the independence from lab-cultivability inherent in “meta-omic” approaches, our approach is substantially faster than current state-of-the-art methodology. We could already identify several novel lipolytic enzymes from a grease- contaminated soil sample using metaproteomics. Through gene synthesis and heterologous expression we can obtain these new biocatalysts in quantity for biochemical characterization.
Some of the biocatalysts we found are lipid degrading enzymes from a restaurant's grease disposal site. In microbial communities from hot springs we found heat- and solvent-stable biocatalysts, that might prove useful in industrial applications. Through collaborations within the DFG-funded Research Training Grant 2341 MiCon we are now using tailor-made substrates to identify enantioselective biocatalysts that could be used for fine-chemical production.